12.1 Worked Example
Get some descriptive stats for the ctDNA data that comes with the package. The nicenames argument is TRUE by default so underscores are replaced by spaces
| n=270 | |
|---|---|
| cohort | |
| A | 50 (19) |
| B | 14 (5) |
| C | 18 (7) |
| D | 88 (33) |
| E | 100 (37) |
| ctdna status | |
| Clearance | 137 (51) |
| No clearance, decrease from baseline | 44 (16) |
| No clearance, increase from baseline | 89 (33) |
| size change | |
| Mean (sd) | -29.7 (52.8) |
| Median (Min,Max) | -32.5 (-100.0, 197.1) |
| Missing | 8 |
12.1.1 set_labels
If we have a lookup table of variable names and labels that we imported from a data dictionary we can set the variable labels for the data frame and these will be used in the rm_ functions
ctDNA_names <- data.frame(var=names(ctDNA),
label=c('Patient ID',
'Study Cohort',
'Change in ctDNA since baseline',
'Number of weeks on treatment',
'Percentage change in tumour measurement'))
ctDNA <- set_labels(ctDNA,ctDNA_names)
rm_covsum(data=ctDNA,
covs=c('cohort','ctdna_status','size_change'))| n=270 | |
|---|---|
| Study Cohort | |
| A | 50 (19) |
| B | 14 (5) |
| C | 18 (7) |
| D | 88 (33) |
| E | 100 (37) |
| Change in ctDNA since baseline | |
| Clearance | 137 (51) |
| No clearance, decrease from baseline | 44 (16) |
| No clearance, increase from baseline | 89 (33) |
| Percentage change in tumour measurement | |
| Mean (sd) | -29.7 (52.8) |
| Median (Min,Max) | -32.5 (-100.0, 197.1) |
| Missing | 8 |
12.1.2 set_var_labels
Individual labels can be changed with with the set_var_labels command
ctDNA <- set_var_labels(ctDNA,
cohort="A new cohort label")
rm_covsum(data=ctDNA,
covs=c('cohort','ctdna_status','size_change'))| n=270 | |
|---|---|
| A new cohort label | |
| A | 50 (19) |
| B | 14 (5) |
| C | 18 (7) |
| D | 88 (33) |
| E | 100 (37) |
| Change in ctDNA since baseline | |
| Clearance | 137 (51) |
| No clearance, decrease from baseline | 44 (16) |
| No clearance, increase from baseline | 89 (33) |
| Percentage change in tumour measurement | |
| Mean (sd) | -29.7 (52.8) |
| Median (Min,Max) | -32.5 (-100.0, 197.1) |
| Missing | 8 |
12.1.3 extract_labels
Extract the variable labels to a data frame
## variable label
## 1 id Patient ID
## 2 cohort A new cohort label
## 3 ctdna_status Change in ctDNA since baseline
## 4 time Number of weeks on treatment
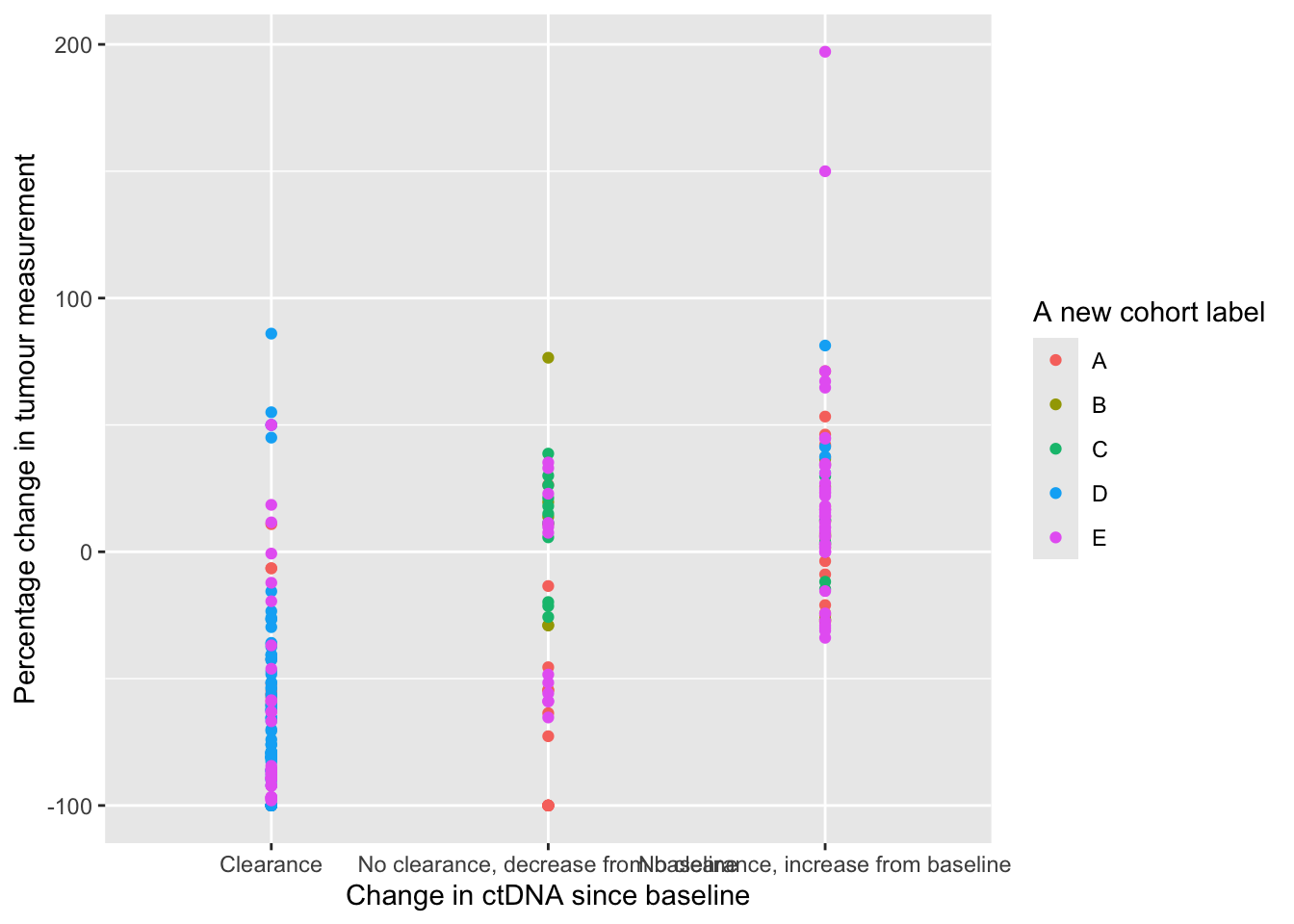
## 5 size_change Percentage change in tumour measurement12.1.4 replace_plot_labels
This function will accept a ggplot plot and replace the variable names (x-axis, y-axis and legend) with the variable labels. This is useful for more professional looking plots.
library(ggplot2)
p <- ggplot(data=ctDNA,aes(x=ctdna_status,y=size_change,colour=cohort))+
geom_point()
replace_plot_labels(p)